NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
New NCBI YouTube video: Create custom databases for BLAST
Friday, March 28, 2014
In the newest NCBI video on YouTube, we show you how to create custom databases in BLAST. This video gives you a step-by-step tutorial on limiting your web BLAST searches to a customized set of sequences.
Come to the NCBI Discovery Workshops on May 6th & 7th!
Friday, March 28, 2014
The NCBI Discovery Workshops will be held on the NIH Campus on May 6 and 7. To get more information and to register, visit the Discovery Workshops homepage.
The NCBI Discovery Workshops, a 2-day event, comprise of four workshops that will teach you how to use the NCBI Web resources more effectively. The May 2014 Workshops consist of four 2.5-hour hands-on sessions, with each session focusing on a different related group of NCBI tools and databases:
Materials from all Discovery Workshops offerings are available from the Education FTP directory.
NCBI will attend the 2014 ACMG Annual Clinical Genetics Meeting
Thursday, March 20, 2014
NCBI staff will attend the 2014 ACMG Annual Clinical Genetics Meeting in Nashville, TN on March 25-29.
At Booth #1105, you’ll be able to do a variety of things including:
- Generate a differential diagnosis based on clinical features,
- Search the ACMG incidental findings gene set against your variant result,
- Join NIH’s open access mission and submit data to GTR and ClinVar,
- And get hands-on help from staff.
For more details, see the GTR page.
NCBI requests feedback on proposed BLAST XML specification update
Monday, March 17, 2014
The BLAST development team is planning to update the BLAST XML specification in the Summer of 2014 and would like feedback from the user community on the proposed changes. This update is designed to improve consistency of the BLAST output with XML standards and implement new and useful elements. The BLAST proposal outlines these intended changes.
If you are a BLAST XML user, please provide feedback on the proposed changes using this web form. We thank you in advance for your input.
RefSeq full release 64 out
Friday, March 14, 2014
The full RefSeq release 64 is now available with nearly 50 million records describing 37,818,139 proteins, 6,198,996 RNAs, and sequences from 33,693 different organisms.
Some important updates include the following:
SNP annotation update: A list of updated organisms and dbSNP annotation summary is available in the SNP RefSeq release notes folder on the FTP site ("refseq63.snp.rpt).
Domain annotation update: RefSeq domain and site features that are provided by the Conserved Domain Database were updated in conjunction with CDD release 3.11. For more information on release 3.11, see the NCBI News story from last month.
New annotation for the updated human reference genome assembly, GRCh38: The Genome Reference Consortium released a major update to the human reference genome assembly (GRCh38) in late December 2013. In January 2014, this updated assembly, plus two other human genome assemblies (HuRef and CHM1_1.1), was annotated using NCBI's eukaryotic genome annotation pipeline which integrated information from curated RefSeqs, cDNAs, ESTs, protein alignments, and RNA-Seq data from the Human BodyMap2 project. Results for all three genomes are available as NCBI Annotation release 106.
More details about the RefSeq release 64 is included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
Orthologous genes and gene regions now accessible through Gene
Wednesday, March 12, 2014
Each Gene record now provides access to orthologous genes and regions in the “General gene information” section of the Gene record (Figure 1). In addition, complex loci in a particular species, such as the human immunoglobulin heavy locus, now have links to the corresponding individual members.
The “Orthologs from Annotation Pipeline” link under the Homology subsection of “General gene information” accesses the set of orthologs in selected vertebrae genomes using the method described in PMCID: PMC3882889. For example, this link from the zebrafish abl1 gene record (Gene ID: 100000720) or from any other member of this orthology group provides 80 orthologous gene records from a wide range of vertebrate species (birds, mammals, turtles, fishes, and the coelacanth). These ortholog data are supplemental to those currently available from the HomoloGene resource also linked under the “Gene information: Homology” section of the Gene record. The Annotation Pipeline method is being improved to include more distantly related organisms in the future.
Region gene records are available for loci that are officially named and are composed of multiple parts or clusters of related genes. The “Related region members” section in a region gene record has a “Review record(s) in Gene” link that provides all genes that are components of the region. For example, the link from the human and mouse immunoglobulin H region records (Gene IDs: 3492 & 11507) provide 182 and 215 records respectively.
The data for both gene orthology groups and gene regions are available in the gene_group.gz file in the Gene area of the NCBI FTP site. In the file, the terms “Ortholog”, “Region members”, and “Region parent” are used to report these new relationships.
New dbGaP online system for registering studies and applying for data access introduces time-saving features
Wednesday, March 12, 2014
In an effort to reduce burden, NIH has developed an online system for researchers and their institutional officials to register studies, submit data, and access data in dbGaP. The online system introduces a number of time-saving features, such as automatically completing data fields from other sources, for example, using eRA Commons to provide the investigator’s name, institution, and Institutional Signing Official.
Tutorials on the online forms for study registration and data access are available on Youtube:
dbGaP: Complete a Study Registration
dbGaP: Renew Authorized Access
dbGaP: Close Out a Controlled Access Project
Additional information can be found in the NIH Guide Notice.
New Sorting and Output Options for E-utilities
Monday, March 10, 2014
E-utilities, the API to the NCBI Entrez system, now offers two new options for data retrieval.
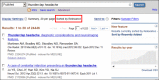
First, ESearch now offers a fully supported &sort parameter that determines the sort order of the UIDs returned. While each Entrez database has a default sort order, each also provides a variety of other sorting options found in the “Display Settings” menu at the top of a search result page. Any of these options may now be given to ESearch using &sort.
Figure
Results sorted by Relevance.
For example, to sort a PubMed retrieval by First Author, simply add “&sort=first+author” to your ESearch URL. If you are also supplying “&usehistory=y”, then the UIDs will also be sorted on the Entrez History so that when retrieved by ESummary or EFetch, the sort order will be retained in that output. Of particular interest is the new relevance sort option now available in PubMed and several other databases. To enable this for ESearch, use “&sort=relevance”.
Second, the output for EInfo, ESearch and ESummary are now available in the popular JSON format. To request data in JSON, simply append “&retmode=json” to the E-utility URL. Click on the following links for example outputs:
Please see the E-utilities documentation for additional details.
- New NCBI YouTube video: Create custom databases for BLAST
- Come to the NCBI Discovery Workshops on May 6th & 7th!
- NCBI will attend the 2014 ACMG Annual Clinical Genetics Meeting
- NCBI requests feedback on proposed BLAST XML specification update
- RefSeq full release 64 out
- Orthologous genes and gene regions now accessible through Gene
- New dbGaP online system for registering studies and applying for data access introduces time-saving features
- New Sorting and Output Options for E-utilities
- NCBI News, March 2014 - NCBI NewsNCBI News, March 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...